Do Proteins Lack Metals, Reflecting Poor Design?
- Casey Luskin
- “Mononuclear iron proteins are oxidoreductases (e.g., aromatic amino acid hydroxylases, aromatic ring cleavage dioxygenases, Fe-superoxide dismutase, lipoxygenases, nitrile hydratase, and Rieske oxygenases), or involved in electron transfer (desulforedoxins, rubredoxins, andphotosynthetic reaction centers).”
- “Hemoproteins are characterized by an iron porphyrin as prosthetic group” and include “oxidoreductases (catalases, peroxidases), electron transfer proteins (cytochromes), or oxygen transport and storage proteins (globins).”
- “Iron–sulfur proteins are characterized by the presence of iron-sulfur clusters containing sulfide-linked di-, tri-, and tetra iron centers in variable oxidation states” and include “ferredoxins, NADH hydrogenases, dehydrogenases, cytochrome creductases, nitrogenases, and other proteins.”
- “Copper proteins are involved in oxygen transport or activation processes and electron transport…” and include “ascorbate oxidase, ceruloplasmin, laccase, nitritereductase, auracyanin, and azurin …”
As I explained in a post yesterday, a TEDx talk from January of this year by MIT bioengineer Erika DeBenedictis, “It’s Time for Intelligent Design,” argues that biology lacks any “intelligent design,” and thus living systems are “imperfect” with “gigantic mistakes,” and we should “play God” in order to “make biology better.” So what are some flaws in biology that we can make “better”? According to Dr. DeBenedictis, “one of the big limitations of biology are the basic building blocks themselves.” Emily Reeves has already addressed Dr. DeBenedictis’s comments about the optimality of amino acids in the genetic code, noting that the set of amino acids used by life is highly optimal. But DeBenedictis goes further and says that the very makeup of the building blocks of life need a major redesign.
“Kind of Boring”
“If we look at the amino acids,” she states, “chemically speaking, they’re actually kind of boring” and “one very easy way to see this is to look at them on a periodic table.” She then shows a periodic table that highlights five chemical elements — hydrogen, carbon, nitrogen, oxygen, and sulfur — she says “Out of all this good stuff, only five chemical elements appear in proteins.” Here’s the slide from the talk:
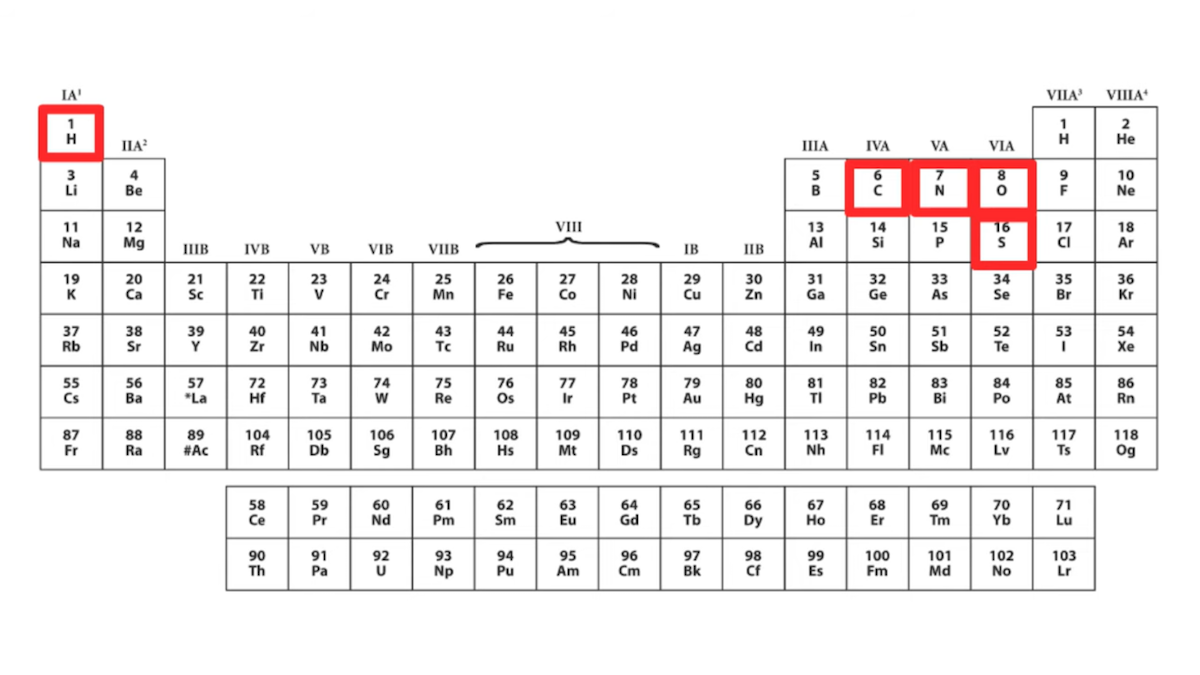
If we are to take Dr. DeBenedictis at her word, then “only five chemical elements appear in proteins,” namely those highlighted in her chart: hydrogen, carbon, nitrogen, oxygen, and sulfur. She further says she wants to create new building blocks and says, “maybe we could get some heavy metals up here.” The message is that proteins only use five elements and that proteins and enzymes essentially ignore the rest of the period table — more evidence of what she calls “imperfect design.”
Dr. DeBenedictis is right that amino acids themselves don’t contain metals. But is it correct for her to state that “only five chemical elements appear in proteins,” suggesting that the rest of the periodic table is absent from protein complexes and not involved in enzyme chemistry? Is it true that proteins don’t make use of metals? No, that is not correct.
Metals, Metals, Everywhere…
Enzymes are proteins that catalyze reactions, and Voet and Voet’s standard textbook Biochemistry states “Nearly one-third of all known enzymes require the presence of metal ions for catalytic activity.” (Voet and Voet 2004, p. 501). The textbook continues:
There are two classes of metal ion-requiring enzymes that are distinguished by the strengths of their ion-protein interactions:
1. Metalloenzymes contain tightly bound metal ions, most commonly transition metal ions such as Fe2+, Fe3+, Cu2+, Zn2+, Mn2+, or Co3+.
2. Metal-activated enzymes loosely bind metal ions from solution, usually the alkali and alkaline earth metal ions Na+, K+, Mg2+, or Ca2+.
Metal ions participate in the catalytic process in three major ways:
1. By binding to substrates so as to orient them properly for reaction.
2. By mediating oxidation-reduction reactions through reversible changes in the metal ion’s oxidation state.
3. By electrostatically stabilizing or shielding negative charges.
VOET AND VOET’S, BIOCHEMISTRY, 2004, P. 501
A 2008 review in Acta Crystallographica Section D, titled “Metals in proteins: correlation between the metal-ion type, coordination number and the amino-acid residues involved in the coordination,” similarly notes how prevalent and important metal usage is in proteins:
Metal ions are constituents of many metalloproteins, in which they have either catalytic (metalloenzymes) or structural functions. … Metals are important for the biological activity of proteins and their removal or the replacement of one metal by another is often accompanied by a loss of or reduction in the biological activity of the protein. Metal ions in proteins can act as structure promoters or can take part in enzymatic reactions. Divalent metal cations such as Zn2+, Mg2+, Cu2+ and Ca2+ are often associated with the catalytic or regulatory activities of proteins that constitute some of the fundamental chemical life processes. For example, Mg in chlorophyll is important for photosynthesis, Cu (together with Fe) has a role in oxygen-carrying proteins and Zn, Mg and Ca can serve as Lewis acids. (emphasis added, internal citations omitted)
A 2011 entry on metalloenzymes in Springer’s Encyclopedia of Geobiology likewise explains that up to a third of known enzymes — i.e., an important type of protein that fosters chemical reactions in cells — use metals as cofactors:
Metalloenzymes are enzyme proteins containing metal ions (metal cofactors), which are directly bound to the protein or to enzyme-bound nonprotein components (prosthetic groups). About one-third of all enzymes known so far are metalloenzymes… metalloenzymes are found in all enzymes families…
The encyclopedia entry goes on to explain:
Besides enzymes, other metalloproteins are involved in non-enzyme electron transfer reactions (cytochromes), may act as storage (e.g., ferritin for iron) or transport proteins (e.g., transferrin for iron). In the latter groups of proteins, the metal storage is reversible and the metal is a temporary component. Also, ribozymes, i.e., RNA molecules with enzyme function may contain structurally and/or functionally important metal ions … and may be therefore termed as metalloenzymes in a broader sense.
Later the entry lists a table summarizing the functions of various metals in enzymes — here’s a condensed version of that table:
| Metallic Element | Roles Played in Enzymes |
| Potassium | Protein stabilization |
| Calcium | Protein stabilization |
| Magnesium | Protein stabilization |
| Iron | Oxygen transport, storage and/or activation; electron transport; superoxide breakdown |
| Manganese | Oxygen release; peroxide and superoxide breakdown |
| Zinc | Protein stabilization; hydrolytic cleavage |
| Copper | Oxygen transport, storage and/or activation; electron transport; superoxide breakdown |
| Molybdenum | Oxygen transfer; N2 activation |
| Cobalt | Free radical reactions; nucleophile |
Going through the entry, it becomes evident that the variety and number of uses of metals in enzymes and proteins are almost endless. Examples listed include:
Heavy Metals
Dr. DeBenedictis’s precise statement in her talk says that we should add “heavy metals” to life. It’s not exactly clear how she would define “heavy metals,” but her slide (see above) and language seem to suggest that proteins don’t use any metals at all. While she is likely trying to simplify things to make a point, it is incorrect to say that proteins don’t use heavy metals. For example, a 2020 paper in Scientific Reports states that many “heavy metals” are “bioavailable,” and these include zinc (Zn), copper (Cu), and nickel (Ni). Multiple sources explain how widely used these heavy metals are in plants.
Zinc: A 2016 study notes how important zinc is in plants:
Zinc (Zn) is an essential micronutrient for plant growth and development. It is also a crucial element for survival of most of the organisms including humans. Zn acts as cofactor of more than 300 proteins, among which majority are zinc finger proteins, RNA polymerases and DNA polymerases. It is the only metal present in all six enzyme classes (oxidoreductase, transferase, hydrolases, lyases, isomerases and ligases). Being a part of structural or catalytic units, Zn regulates the activities, conformational stabilization and folding of various proteins (enzymes). In addition to this, role of Zn in membrane integrity and stabilization, alleviation of oxidative stress — and as intracellular second messenger has also been reported. Zn is involved in a number of plant physiological processes such as hormone regulation (e.g. tryptophan synthesis, a precursor of IAA), signal transduction via mitogen activated protein kinases, repair processes of PS II complex during photoinhibition and maintenance of CO2 concentration in mesophyll. Peck and McDonald (2010) — confirms the participation of Zn in regulation of Rubisco activity along with alleviation from adverse effects of heat stress in wheat. Thus, Zn essentiality for plant system is illustrated from literature cited above.” [Internal citations removed.]
According to a horticulture website:
Zinc activates enzymes that are responsible for the synthesis of certain proteins. It is used in the formation of chlorophyll and some carbohydrates, conversion of starches to sugars and its presence in plant tissue helps the plant to withstand cold temperatures. Zinc is essential in the formation of auxins, which help with growth regulation and stem elongation.
Zinc plays similar roles in animal biochemistry.
Copper: A 2018 paper notes that “copper is an essential element for proper growth and development of plants. Copper in plants is functioning as a catalyst in respiration and photosynthesis. It is vital element for the creation of lignin in plant cell walls and in the case of enzymes responsible for protein synthesis. It also influences the disease resistance and reproduction.” The horticulture site likewise explains the role of copper in plants:
Copper activates some enzymes in plants which are involved in lignin synthesis and it is essential in several enzyme systems. It is also required in the process of photosynthesis, is essential in plant respiration and assists in plant metabolism of carbohydrates and proteins. Copper also serves to intensify flavor and color in vegetables and color in flowers.
Nickel: As for nickel, the same horticulture website explains:
Nickel is a component of some plant enzymes, most notably urease, which metabolizes urea nitrogen into useable ammonia within the plant. Without nickel, toxic levels of urea can accumulate within the tissue forming necrotic legions on the leaf tips. In this case, nickel deficiency causes urea toxicity. Nickel is also used as a catalyst in enzymes used to help legumes fix nitrogen. There is evidence that nickel helps with disease tolerance in plants, although it is still unclear how this happens.
Ideas Have Consequences
I could go on, citing review papers, research papers, biochemistry textbooks, and other authorities explaining how important metals are to the basic functions of numerous proteins. At the very least, it seems an oversight to criticize biology for not using “heavy metals” and yet fail to acknowledge any of the numerous instances where metals are used to fulfill core functions in enzymes and other proteins. To show a full periodic table that highlights only hydrogen, carbon, nitrogen, oxygen, and sulfur, and then to say “Out of all this good stuff, only five chemical elements appear in proteins,” severely underrepresents the degree to which metals are used in biology.
I infer no bad faith or ignorance on Dr. DeBenedictis’s part. Although her Covid-lockdown-era TEDx talk was recorded privately, with opportunities for multiple takes to get things right, I believe what she probably meant was that amino acids don’t use metals, and it was a simple misstatement. But there’s still a lesson here.
Ideas have consequences. When we adopt a materialistic perspective which assumes that biology has no intelligent design, and instead contains fundamental flaws or “gigantic mistakes,” we’ll be quicker to assume that features of biological systems exist for no good reason. This discourages us from investigating why things are the way they are. We’ll be more likely to miss good design features in biology, which is exactly what I think has happened here.
Materialism’s blind spots often exist right where important biological features are found. We’ve certainly seen this blind spot in past thinking about “junk DNA,” where a standard evolutionary framework discouraged research into the functions of this DNA. Today, we know “junk DNA” has key biological functions. But materialistic thinking doesn’t encourage us to understand the design principles that underly biology.
Intelligent design is an idea that has consequences, too. And when it comes to understanding biology, ID is a paradigm that can lead to fruitful research predictions. Those predictions start with the suspicion that there’s probably a good reason why things are the way they are, and task is up to us to figure out what those reasons are.