Computer Software Sheds Light on Human and Chimp DNA Similarity
Walter Myers III
Walter Myers III
Recently I had the opportunity to hear Discovery Institute’s Stephen Meyer provide an update on the progress and current state of the theory of intelligent design. At the end of Meyer’s lecture, he took questions from the audience. Inevitably, the question came up about humans and chimpanzees with their “98 percent” similarity in DNA. Isn’t that evidence in favor of evolution and against design?
Meyer’s reply was to compare DNA code to differing computer programs that share an underlying code base. As a professional software engineer over more decades than I care to express here, I can attest to the accuracy of Meyer’s comparison. As he demonstrated in his book Signature in the Cell, the cell is a microscopic factory bustling with the activity of thousands of tiny machines built from the instructions provided by DNA code in the nucleus of the cell.
I am not going to enter into the debate about what precisely is the percentage of similarity in DNA between humans and chimps. That is wholly immaterial to the point I want to make. Instead, let’s see how the analogy that Meyer presented holds up by providing more depth and color using a practical example from the hardware and software most of us use in everyday life. Everyone reading this post (I suspect) is using a browser on either a computer, tablet, or smartphone. The device you are using has something installed on it called an operating system (OS). That is defined as “the collection of software that directs a computer’s operations, controlling and scheduling the execution of other programs, and managing storage, input/output, and communication resources.” The operating system provides all of the underlying functions necessary for the browser or any other application software (program) you may access on your device. Whatever the device you are using, the OS consists of tens of millions of lines of code. For example, the Windows operating system is estimated to have in excess of 50 million lines of code.
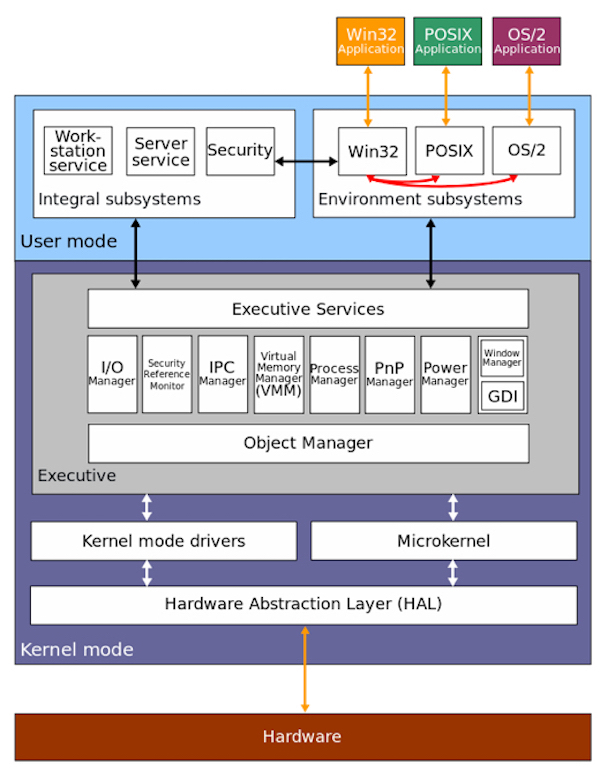
The diagram below represents the architecture of Windows NT, which is the line of operating systems produced and sold by Microsoft. Actually this diagram is a bit dated as Windows has “evolved” quite a bit with new features since 2000, but the fundamental concepts have not changed. It’s not essential that you understand this in full, but note the various subsystems that make up a modern OS.
Specifically, you have the “kernel” which is the core of the OS connecting application software to the underlying hardware. The kernel exercises complete control over the system and is fully protected from user applications, providing a set of well-defined interfaces by which an application can interact with the underlying services. You can liken the kernel here to the nucleus of a cell, which maintains the security of DNA and controls the functions of the entire cell by regulating gene expression. On top of the kernel, you have a “user” mode coordinating with the kernel that provides higher-level services such as your user interface, authentication mechanism (for logging in), and the environment in which your application code runs (in this case, your browser). This would be analogous to the working proteins in the cell which would be the running code performing the everyday work of the cell.

Now let’s focus further on application code. While the OS is code that an OS company writes, such as Microsoft, Apple, or various open-source Linux distribution companies, applications are written by software developers. Applications themselves can also run to millions of lines of code. It depends upon the complexity and functionality of the application itself. What software developers have discovered over the decades, however, is that there are specific functions or patterns that developers perform over and over, and thus a considerable part of the software business consists of “third-party” developers writing and selling reusable “libraries” that make work easier for other developers. For example, in a typical application, you might have a library that assists with building the user interface, a library for database access, or a library for communications over a wireless network.
Applications on a smartphone, such as Facebook, Instagram, or Snapchat, can be thousands of lines of code, accessing component libraries that provide services made up of thousands or millions of lines of code, and of course accessing the aforementioned millions of lines of code in the underlying OS.
Now, comparing this to humans and chimps, what do we find? While much of the DNA code may be the same, the parts that are not the same have significant differences. The programs I described above, such as Facebook, Instagram, or Snapchat, have different purposes, yet they all depend on the same OS that consists of tens of millions of lines of code. To be specific, let’s say you are using an iPhone with iOS 11 (the Apple mobile OS) installed. iOS is estimated to take up about 4 GB of space on your iPhone. Facebook takes up about 297 MB. Snapchat is about 137 MB. Instagram is about 85 MB. Respectively, that’s 7.4 percent, 3.4 percent, and 2.1 percent of the size of iOS. Now would anyone say that Facebook, Instagram, and Snapchat are pretty much the same thing since they are each well over 90 percent the same? Of course not. It’s not so different with humans and chimps. In the case of these programs, the vast majority of their total code base is shared, yet each is a distinct creative expression that leverages a shared base of code. In the case of humans and chimps, one would expect a designer to use shared code where functions are the same, and different (new) code where functions are different. When we examine computer programs, which are the inventions of human minds, why would they not reflect the mind of the designer that wrote the code to produce humans, chimps, and every other biological organism?
There is a further relevant analogy between application software and DNA code. In biological organisms, not all genes are expressed in every cell. For example, there are specific genes active in liver cells, specific genes active in heart muscle cells, and specific genes active in brain cells. Different cell types express themselves in both appearance and function. So not all of the DNA code is in use in each cell. Additionally, environmental factors affect what genes are expressed in a group of cells, allowing an organism to respond in various ways to the situations in which it finds itself. Similarly, with software programs, not all pathways to all code are in use. There are “settings,” whether set by the user or programmed automatically in the application by the developer, that determine how a program will individually function. For example, when a user changes the privacy, language, or chat settings in the Facebook mobile app, it modifies the many pathways the code may execute. Or if a malevolent user tries to log in to a program multiple times, attempting to hack into a user account, the program will itself execute a code pathway to lock the malevolent user out and notify the legitimate user.
Again, the functions in a computer program reflect the mind of the human designer. In the same way, the functions in the human being programming a computer reflect the mind of the designer of both humans and chimps.